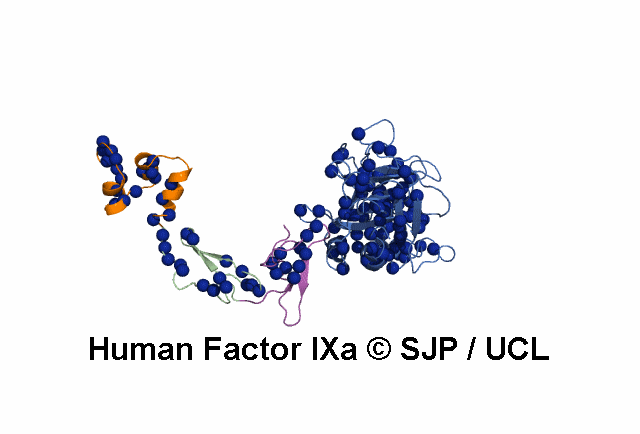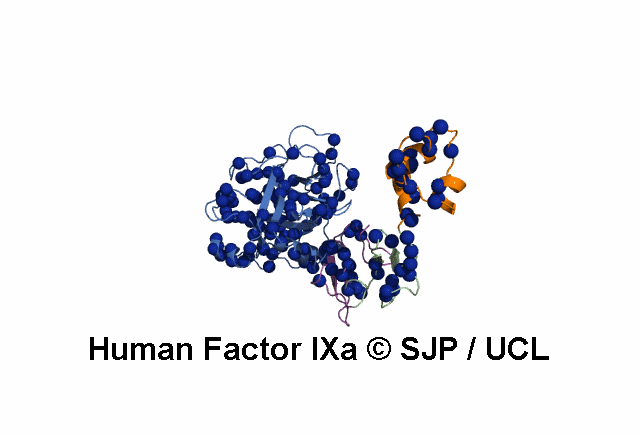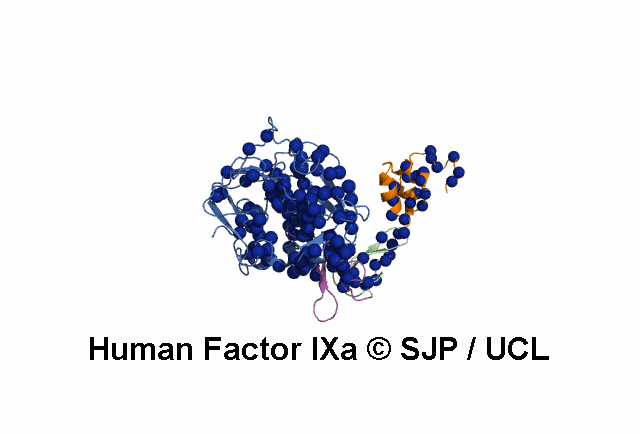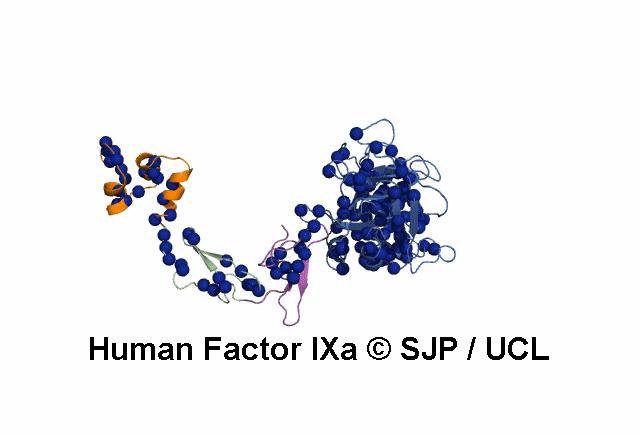
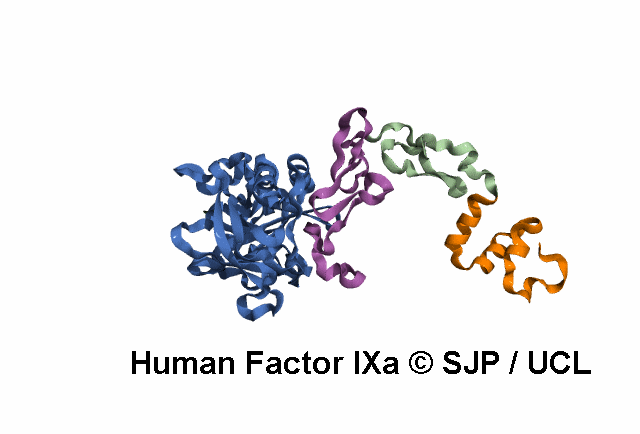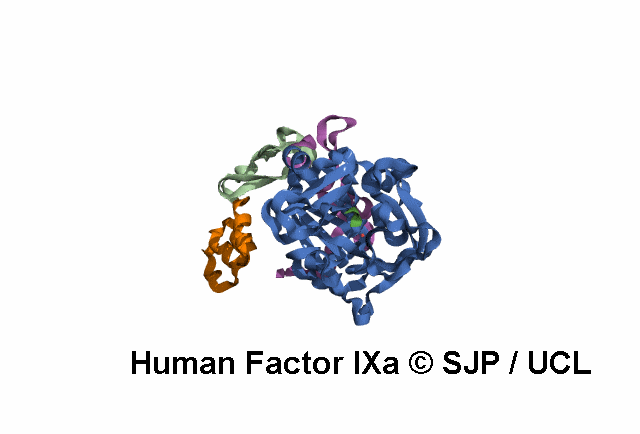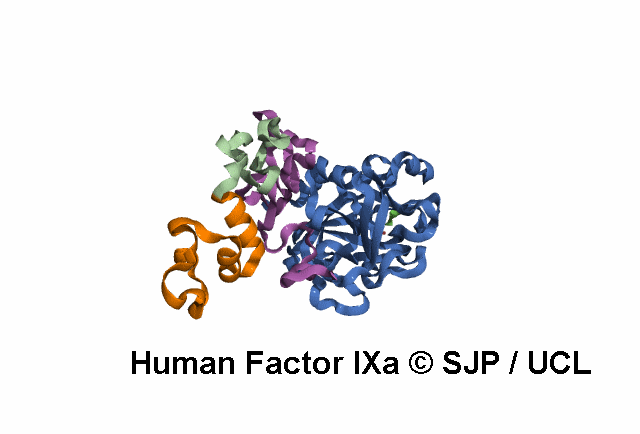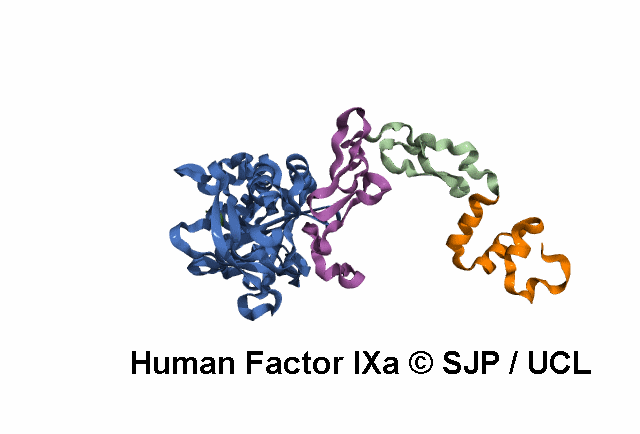
Point Mutations Mapped on to the FIX sequence
- All amino acids that are colored in red-bold font have associated mutations (point) within the database.
- More information on each amino acid mutation and its effects on the structure can be gained by clicking it.
- This display is automatically generated and represents all the amino acid mutations at that residue within FIX.
KEY = DOMAINS:: GLA -- EGF1 -- EGF2 -- Serine Protease
MQRVNMIMAE SPGLITICLL GYLLSAECTV FLDHENANKI LNRPKRYNSG 50
KLEEFVQGNL ERECMEEKCS FEEAREVFEN TERTTEFWKQ YVDGDQCESN 100
PCLNGGSCKD DINSYECWCP FGFEGKNCEL DVTCNIKNGR CEQFCKNSAD 150
NKVVCSCTEG YRLAENQKSC EPAVPFPCGR VSVSQTSKLT RAETVFPDVD 200
YVNSTEAETI LDNITQSTQS FNDFTRVVGG EDAKPGQFPW QVVLNGKVDA 250
FCGGSIVNEK WIVTAAHCVE TGVKITVVAG EHNIEETEHT EQKRNVIRII 300
PHHNYNAAIN KYNHDIALLE LDEPLVLNSY VTPICIADKE YTNIFLKFGS 350
GYVSGWGRVF HKGRSALVLQ YLRVPLVDRA TCLRSTKFTI YNNMFCAGFH 400
EGGRDSCQGD SGGPHVTEVE GTSFLTGIIS WGEECAMKGK YGIYTKVSRY 450
VNWIKEKTKL T
KLEEFVQGNL ERECMEEKCS FEEAREVFEN TERTTEFWKQ YVDGDQCESN 100
PCLNGGSCKD DINSYECWCP FGFEGKNCEL DVTCNIKNGR CEQFCKNSAD 150
NKVVCSCTEG YRLAENQKSC EPAVPFPCGR VSVSQTSKLT RAETVFPDVD 200
YVNSTEAETI LDNITQSTQS FNDFTRVVGG EDAKPGQFPW QVVLNGKVDA 250
FCGGSIVNEK WIVTAAHCVE TGVKITVVAG EHNIEETEHT EQKRNVIRII 300
PHHNYNAAIN KYNHDIALLE LDEPLVLNSY VTPICIADKE YTNIFLKFGS 350
GYVSGWGRVF HKGRSALVLQ YLRVPLVDRA TCLRSTKFTI YNNMFCAGFH 400
EGGRDSCQGD SGGPHVTEVE GTSFLTGIIS WGEECAMKGK YGIYTKVSRY 450
VNWIKEKTKL T